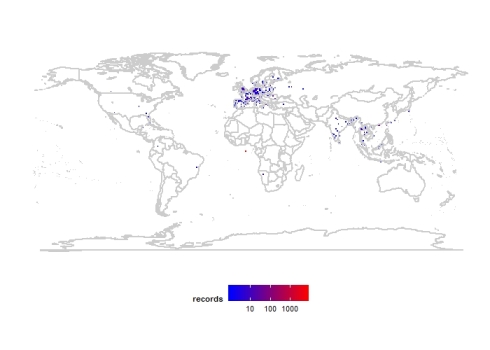
Guest post by Qingyue Xu
Compiling taxonomic checklists from varied sources of data is a common task that biodiversity informaticians encounter. In the GSoC 2017 project Parser for Biodiversity checklists, my overall goal is to extract taxonomic names from given text into a tabular format so that easy aggregation of biodiversity data in a structured format that can be used for further processing can be facilitated.
I mainly plan to build three major functions which serve different purposes and take various sources of text into account.
However, before building functions, we need to first identify and cover as many different formats of scientific names as possible. The inconsistencies of scientific names make things complicated for us. The common rules for scientific names follow the order of:
genus, [species], [subspecies], [author, year], [location]
Many components are optional, and some components like author and location can be one or even more. Therefore, when we’re parsing the text, we need to analyze the structure of the text and match it with all possible patterns of scientific names and identify the most likely one. To resolve the problem more accurately, we can even draw help from NLTK (Natural Language Toolkit) packages to help us identify “PERSON” and “LOCATION” so that we can analyze the components of scientific names more efficiently.
Function: find_taxoname (url/input_file, output_file)
- Objective: This is a function to search scientific names with supplied texts, especially applied to the situation when the text is not well structured.
- Parameters: The first parameter is the URL of a web page (HTML based) or the file path of a PDF/TXT file, which is our source text to search for the biology taxonomic names. The second parameter is the file path of the output file and it will be in a tabular format including columns of the genus, species, subspecies, author, year.
- Approach: Since this function is intended for the unstructured text, we can’t find a certain pattern to parse the taxonomic names. What we can do is utilizing existing standard dictionaries of scientific names to locate the genus, species, subspecies. By analyzing the surrounding structure and patterns, we can find corresponding genus, species, subspecies, author, year, if they exist, and output the findings in a tabular format.
Function: parse_taxolist(input_file, filetype, sep, output_file, location)
- Objective: This is a function to parse and extract taxonomic names from a given structured text file and each row must contain exactly one entry of the scientific names. If the location information is given, the function can also list and return the exact location (latitude and longitude) of the species. The output is also in a tabular format including columns of genus, species, subspecies, author(s), year(s), location(s), latitude(s), longitude(s).
- Parameters: The first parameter is the file path of the input file and the second parameter is the file type, which supports txt, PDF, CSV types of files. The third parameter ‘sep’ should indicate the separators used in the input file to separate every word in the same row. The fourth parameter is the intended file path of the output file. The last parameter is a Boolean, indicating whether the input file contains the location information. If ‘true’, then the output will contain detailed location information.
- Approach: The function will parse the input file based on rows and the given separators into a well-organized tabular format. An extended function is to point the exact location of the species if the related information is given. With the location info such as “Mirik, West Bengal, India”, the function will return the exact latitude and longitude of this location as 26°53’7.07″N and 88°10’58.06″E. It can be realized through crawling the web page of https://www.distancesto.com/coordinates or utilizing the API of Google Map. This is also a possible solution to help us identify whether the content of the text represents a location. If it cannot get exact latitude and longitude, then it’s not a location. If a scientific name doesn’t contain location information, the function will return NULL value for the location part. If it contains multiple locations, the function will return multiple values as a list as well as the latitudes and longitudes.
Function: recursive_crawler(url, htmlnode_taxo, htmlnode_next, num, output_file, location)
- Objective: This function is intended to crawl the web pages containing information about taxonomic names recursively. The start URL must be given and the html_node of the scientific names should also be indicated. Also, if the text contains location info, the output will also include the detailed latitude and longitude.
- Parameters: The first parameter is the start URL of the web page and the following web pages must follow the same structure as the first web page. The second parameter is the html_node of the taxonomic names, such as “.SP .SN > li”. (There’re a lot of tools for the users to identify the HTML nodes code for certain contexts). The third parameter is the html_node of the next page, which can lead us to the next page of another genus. The fourth parameter ‘num’ is the intended number of web pages the user indicates. If ‘num’ is not given, the function will automatically crawl and stop until the htmlnode_next cannot return a valid URL. The next two parameters are the same with the above two functions.
- Approach: For the parsing part and getting the location parts, the approach is the same as the above functions. For the crawling part, for a series of structured web pages, we can parse and get valid scientific names based on the given HTML nodes. The HTML nodes for the next pages should also be given, and we can always get the URL of the next page by extracting it from the source code. For example, the following screenshot from the web page we used provides a link which leads us to the next page. By recursively fetching the info from the current page and jump to the following pages, we can output a well-organized tabular file including all the following web pages.
- Other possible functionalities to be realized
Since inconsistencies might exist in the format of scientific names, I also need to construct a function to normalize the names. The complication always lies in the author part, and there can be two approaches to address the problem. The first one is still analyzing the structure of the scientific name and we can try to capture as many exceptions as possible, such as author names which have multiple parts or there’re two authors. The second approach is to draw help from the NLTK package to identify possible PERSON names. However, when it gets too complicated, the parsing result won’t be very accurate all the time. Therefore, we can add a parameter to suggest how reliable our result is and indicate the need for further manual process if the parser cannot work reliably.